Examples of workflows¶
Example #1¶
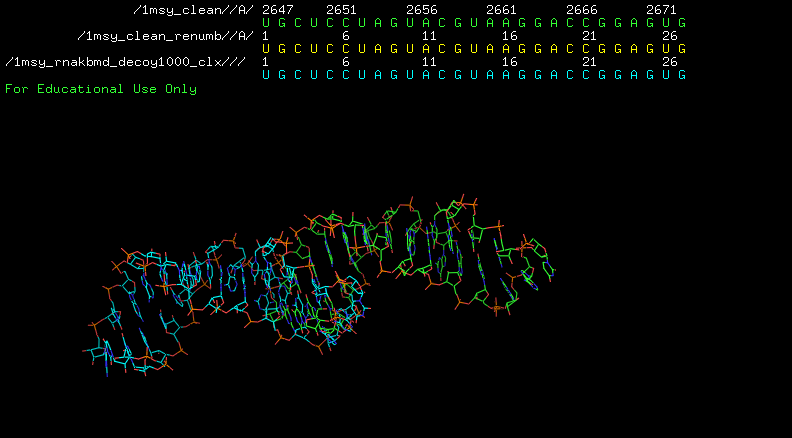
The native:
[mq] md_1msy_clx$ cat 1msy_clean.pdb.outCR
Classifier: Clarna
chains: A 2647 2673
A 2648 A 2672 bp G U WW_cis 0.8732
A 2649 A 2671 bp C G WW_cis 0.9160
A 2650 A 2670 bp U A WW_cis 0.9289
A 2651 A 2669 bp C G WW_cis 0.9439
A 2652 A 2668 bp C G WW_cis 0.9281
A 2655 A 2656 bp G U SH_cis 0.9227
A 2656 A 2665 bp U A WH_tran 0.8526
A 2657 A 2664 bp A G HS_tran 0.8513
A 2658 A 2663 bp C G WW_cis 0.9421
A 2659 A 2662 bp G A SH_tran 0.7619
but analyzed structres are like:
[mq] md_1msy_clx$ cat struc/1msy_rnakbmd_decoy1478_clx.pdb.outCR
Classifier: Clarna
chains: A 1 27
2 26 bp G U WW_cis 0.7196
3 25 bp C G WW_cis 0.6702
4 24 bp U A WW_cis 0.8911
5 23 bp C G WW_cis 0.8925
6 22 bp C G WW_cis 0.9026
9 10 bp G U SH_cis 0.8714
10 19 bp U A WH_tran 0.7279
11 18 bp A G HS_tran 0.8810
12 17 bp C G WW_cis 0.9115
You have to renumber 1msy_clean.pdb to 1:27:
$ rna_pdb_tools.py --edit 'A:2647-2673>A:1:17' 1msy_clean.pdb > 1msy_clean_renumb.pdb
Example #2¶
Listing:
$ rna_pdb_tools.py --get-seq 1nuj_rnakbmd_decoy1000_clx.pdb
> 1nuj_rnakbmd_decoy1000_clx.pdb A:1-13
CGGACCGAGCCAG
> 1nuj_rnakbmd_decoy1000_clx.pdb B:14-24
GCUGGGAGUCC
$ rna_pdb_tools.py --get-seq 1nuj_clean.pdb
> 1nuj_clean.pdb A:18-30
CGGACCGAGCCAG
> 1nuj_clean.pdb B:39-49
GCUGGGAGUCC
$ rna_pdb_tools.py --edit 'A:18-30>A:1-13,B:39-49>B:14-24' 1nuj_clean.pdb > 1nuj_clean_renumber.pdb
$ rna_pdb_tools.py --get-seq 1nuj_clean_renumber.pdb
> 1nuj_clean_renumber.pdb A:1-13
CGGACCGAGCCAG
> 1nuj_clean_renumber.pdb B:14-24
GCUGGGAGUCC
Example #3¶
Starting structure doesn’t have chain id:
# add chain A
$ parallel "rna_add_chain.py -c A {} > ../struc_with_chain/{}" ::: *.pdb
# edit the second part of the new chain A as B
$ parallel "rna_pdb_gtools.py --edit 'A:14-27>B:14-27' {} > out/{}" ::: *.pdb
Example #4 Calculate RMSDs of unstandardized structures (RNA Puzzle #1)¶
You try to calculate RMSDs for RNA Puzzles #1:
$ rna_calc_rmsd.py -t 1_solution_0_rpr.pdb *.pdb
method: all-atom-built-in
# of models: 15
1_bujnicki_1_rpr.pdb 5.71 978
1_bujnicki_2_rpr.pdb 6.16 978
1_bujnicki_3_rpr.pdb 5.3 978
1_bujnicki_4_rpr.pdb 4.95 978
1_bujnicki_5_rpr.pdb 5.1 978
Error: # of atoms is not equal target (1_solution_0_rpr.pdb):978 vs model (1_chen_1_rpr.pdb):975
you can see that there is a different number of atoms in 1_solution_0_rpr.pdb and 1_chen_1_rpr.pdb.
To see more you can run diffpdb.
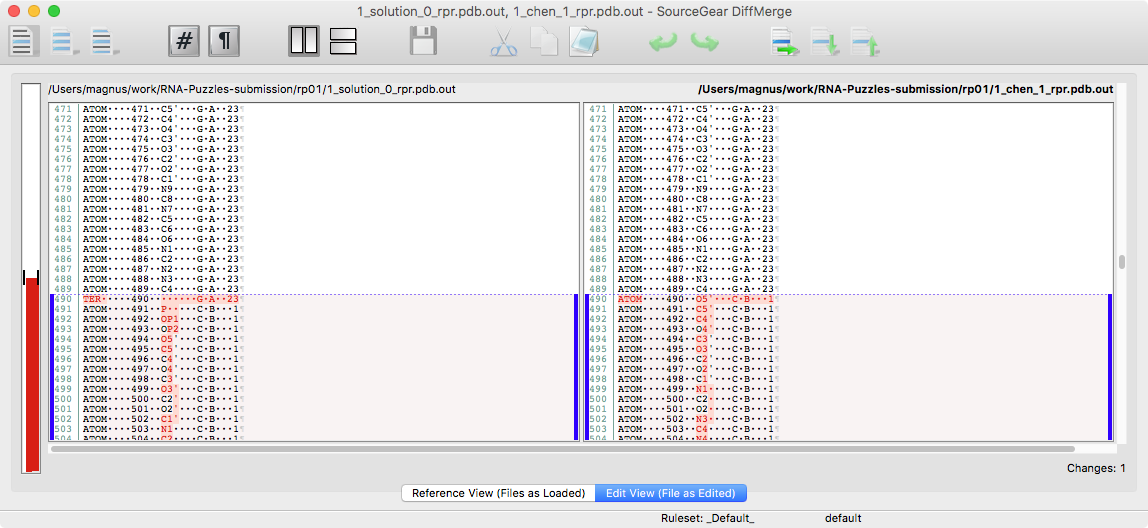
you see that something is wrong. To fix it, run:
$ rna_pdb_tools.py --rpr --inplace *.pdb
93% (15 of 16) |######################################################################################################################## | Elapsed Time: 0:00:03 ETA: 0:00:00
you can tail the files:
$ tail *.pdb
==> 1_bujnicki_1_rpr.pdb <==
ATOM 971 N7 G B 23 -16.558 -3.375 78.345 1.00 0.00 N
ATOM 972 C5 G B 23 -17.169 -2.575 77.384 1.00 0.00 C
ATOM 973 C6 G B 23 -17.589 -2.874 76.053 1.00 0.00 C
ATOM 974 O6 G B 23 -17.497 -3.930 75.430 1.00 0.00 O
ATOM 975 N1 G B 23 -18.234 -1.800 75.459 1.00 0.00 N
ATOM 976 C2 G B 23 -18.441 -0.576 76.049 1.00 0.00 C
ATOM 977 N2 G B 23 -19.127 0.345 75.382 1.00 0.00 N
ATOM 978 N3 G B 23 -18.053 -0.282 77.292 1.00 0.00 N
ATOM 979 C4 G B 23 -17.419 -1.324 77.898 1.00 0.00 C
...
==> 1_chen_1_rpr.pdb <==
ATOM 971 N7 G B 23 -14.462 -1.101 79.998 1.00 0.00 N
ATOM 972 C5 G B 23 -14.952 -0.485 78.839 1.00 0.00 C
ATOM 973 C6 G B 23 -15.577 -1.020 77.655 1.00 0.00 C
ATOM 974 O6 G B 23 -15.822 -2.189 77.351 1.00 0.00 O
ATOM 975 N1 G B 23 -15.972 -0.051 76.763 1.00 0.00 N
ATOM 976 C2 G B 23 -15.787 1.274 76.944 1.00 0.00 C
ATOM 977 N2 G B 23 -16.269 2.059 76.021 1.00 0.00 N
ATOM 978 N3 G B 23 -15.224 1.822 78.022 1.00 0.00 N
ATOM 979 C4 G B 23 -14.818 0.884 78.935 1.00 0.00 C
TER 980 G B 23
==> 1_solution_0_rpr.pdb <==
ATOM 971 N7 G B 23 22.256 -1.292 27.403 1.00 34.10 N
ATOM 972 C5 G B 23 22.625 -0.176 28.135 1.00 31.12 C
ATOM 973 C6 G B 23 23.470 -0.096 29.260 1.00 28.80 C
ATOM 974 O6 G B 23 24.062 -1.036 29.804 1.00 28.26 O
ATOM 975 N1 G B 23 23.616 1.224 29.705 1.00 27.28 N
ATOM 976 C2 G B 23 22.971 2.318 29.112 1.00 28.31 C
ATOM 977 N2 G B 23 23.179 3.538 29.655 1.00 27.03 N
ATOM 978 N3 G B 23 22.170 2.245 28.047 1.00 28.85 N
ATOM 979 C4 G B 23 22.041 0.961 27.632 1.00 28.58 C
TER 980 G B 23%
so now you can see that the files look the same. Let’s try to calculate RMSDs again:
$ rna_calc_rmsd.py -t 1_solution_0_rpr.pdb *.pdb
method: all-atom-built-in
# of models: 16
1_bujnicki_1_rpr.pdb 5.71 978
1_bujnicki_2_rpr.pdb 6.16 978
1_bujnicki_3_rpr.pdb 5.3 978
1_bujnicki_4_rpr.pdb 4.95 978
1_bujnicki_5_rpr.pdb 5.1 978
1_chen_1_rpr.pdb 4.35 978
1_chen_1_rpr_v2.pdb 4.35 978
1_das_1_rpr.pdb 3.97 978
1_das_2_rpr.pdb 4.48 978
1_das_3_rpr.pdb 3.43 978
1_das_4_rpr.pdb 3.92 978
1_das_5_rpr.pdb 4.57 978
1_dokholyan_1_rpr.pdb 7.25 978
1_major_1_rpr.pdb 4.34 978
1_santalucia_1_rpr.pdb 5.76 978
1_solution_0_rpr.pdb 0.0 978
# of atoms used: 978
csv was created! rmsds.csv
worked! :-)
This is a real-life case, https://github.com/mmagnus/RNA-Puzzles-Normalized-submissions/tree/master/rp01.